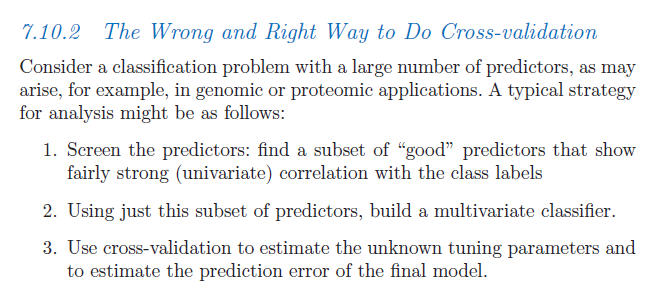
Cross-validation (CV) is nowadays being widely used for model assessment in predictive analytics tasks; nevertheless, cases where it is incorrectly applied are not uncommon, especially when the predictive model building includes a feature selection stage.
I was reminded of such a situation while reading this recent Revolution Analytics blog post, where CV is used to assess both the feature selection process (using genetic algorithms) and the final model selection using the features previously selected. In summary, the procedure followed in the above post is:
-
- Select a number of “good” predictors, using the genetic algorithms (GA) method provided by the caret R package
-
- Using just this subset of predictors, build an SVM classifier
- Use cross-validation to estimate the unknown tuning parameters of the classifier, and to estimate the prediction error of the final model.
And we should be fine. Right?
Well, no…!
The above 3-step procedure vividly illustrates a recurring mistake when applying CV for model assessment with a feature selection stage in-between; consider the following excerpt from The Elements of Statistical Learning (Hastie et al., 2009):
The very title of the section above indeed suggests that there are some common “traps” when applying CV; Hastie et al. proceed to ask, Is this a correct application of cross-validation? And the answer turns out to be: no.
Why is that? Before delving into the statistical arguments, Hastie et al. provide an intuitive explanation:
As Hastie et al. explain in detail, the correct way in this case is to apply feature selection inside each one of the CV folds; you can watch a short video on the topic (“Cross-validation: right and wrong“) from their Statistical Learning MOOC (highly recommended), as well as a couple of relevant slides they have put together here.
Possibly the most highly cited reference on the issue, which leads to what we call selection bias, is a 2002 paper by Ambroise & McLachlan in the Proceedings of the National Academy of Sciences of the USA (open access – emphasis ours):
As explained above, the CV error of the prediction rule R obtained during the selection of the genes provides a too-optimistic estimate of the prediction error rate of R. To correct for this selection bias, it is essential that cross-validation or the bootstrap be used external to the gene-selection process. […]
In the present context where feature selection is used in training the prediction rule R from the full training set, the same feature-selection method must be implemented in training the rule on the M − 1 subsets combined at each stage of an (external) cross-validation of R for the selected subset of genes.
The issue has been discussed several times since in the academic literature, with identical conclusions; see for example a 2006 paper by Varma & Simon in BMC Bioinformatics (open access):
However, CV methods are proven to be unbiased only if all the various aspects of classifier training takes place inside the CV loop. This means that all aspects of training a classifier e.g. feature selection, classifier type selection and classifier parameter tuning takes place on the data not left out during each CV loop. It has been shown that violating this principle in some ways can result in very biased estimates of the true error. One way is to use all of the training data to choose the genes that discriminate between the two classes and only change the classifier parameters inside the CV loop. This violates the principle that feature selection must be done for each loop separately, on the data that is not left out. As pointed out by Simon et al. [2], Ambroise and McLachlan [3] and Reunanen [4], this gives a very biased estimate of the true error; not much better than the resubstitution estimate. Over-optimistic estimates of error close to zero are obtained, even for data where there is no real difference between the two classes.
Notice that the fact that in Step 1 of the above mentioned Revolution Analytics blog post the features themselves are selected via a separate CV procedure is irrelevant to the argument; essentially, “use all of the training data to choose the genes that discriminate between the two classes and only change the classifier parameters inside the CV loop” is exactly what is performed in that post, which clearly “violates the principle that feature selection must be done for each loop separately“.
Even for people that do not frequent academic publication sites, the issue receives a whole section in the (highly recommended) Applied Predictive Modeling book (Section 19.5 Selection Bias), as well as in the extensive online documentation of the R package caret (“Resampling and External Validation“). For a practical account of the consequences that such a mistaken application of CV can incur, see this post, which was reposted in the Kaggle blog with the characteristic title “The Dangers of Overfitting or How to Drop 50 spots in 1 minute“. Quoting:
as the competition went on, I began to use much more feature selection and preprocessing. However, I made the classic mistake in my cross-validation method by not including this in the cross-validation folds (for more on this mistake, see this short description or section 7.10.2 in The Elements of Statistical Learning). This lead to increasingly optimistic cross-validation estimates.
[…]
Lessons learned
[…]
- On a related note, perform cross-validation the right way: include all training (feature selection, preprocessing, etc.) in each fold.
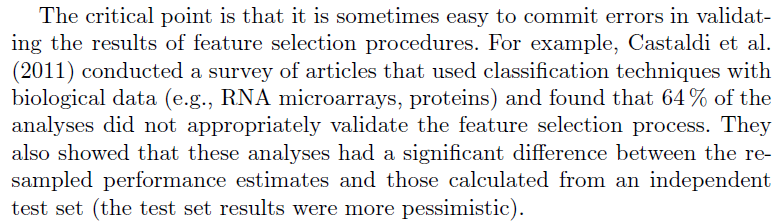
Unfortunately, as we mentioned in the beginning, the issue is far from uncommon among both academics and practitioners, especially when a feature selection procedure is involved; quoting from Applied Predictive Modelling (you can find the referenced paper by Castaldi et al. here):
Or from The Elements of Statistical Learning:
So, the bottom line here is: if you are using feature selection in your data processing pipeline, you have to ensure that it is included in the CV (or whatever resampling technique you use) for your model assessment; otherwise, your results will be biased, and your model expected performance will be worse than assessed.
There is one exception to the above rule, and that is when your feature selection process is unsupervised, i.e. it does not take into account the response variable; quoting again from The Elements of Statistical Learning:
Nevertheless, in practice this procedure is normally performed at the data preprocessing stage, and it is not considered part of feature selection proper.
- Streaming data from Raspberry Pi to Oracle NoSQL via Node-RED - February 13, 2017
- Dynamically switch Keras backend in Jupyter notebooks - January 10, 2017
- sparklyr: a test drive on YARN - November 7, 2016




Dear Mr.
First, many thanks for your useful comment about this article (http://blog.revolutionanalytics.com/2015/12/caret-genetic.html). Then, I would appreciate if you could kindly let me know how to modify that code.
Best regards,
Thank you sir. It would be of great help to me if you tell me how to modify that code.
Thanks.
Dear Christos, Thanks for posting good and informative article of CV and feature selection. This article is based on the “Feature with caret’s Genetic Algorithm Option” article iN which as per you no CV has been used for feature selection, but I wonder about it as ga_ctrl <- gafsControl(functions = rfGA, # Assess fitness with RF method = "cv", # 10 fold cross validation genParallel=TRUE, # Use parallel programming allowParallel = TRUE is show the the feature have been selected for 10 CV folds. However, I am new to this field, I just need your help to modify this code… Read more »
I find it helpfull and very intersting article and i appreciate your efficient explanation. Here we have to take into consideration to correct selection bias So we have to use external corss validation rather than internal cross validation which means we have to apply tuning parameters and feature selection separately in each fold and not in the full training set
Bravo. Thx a lot. still few doubts on how to use the PCA in this context , but at least it is clear that I made a mistake. thx a lot!
[…] How NOT to perform feature selection! […]
[…] How NOT to perform feature selection!. […]
[…] the correct & wrong way to perform such processes; I have summarized the issue in a blog post, How NOT to perform feature selection! – and although the discussion is about cross-validation, it can be easily seen that the […]
[…] https://www.nodalpoint.com/not-perform-feature-selection/ […]