I was very happy to find out that, in the latest version (4.2.1) of Oracle Big Data Lite VM, all the R-related issues I had located and reported in the past (see here and here) have been resolved. Nevertheless, some new issues have emerged. Below are my findings and workarounds (if you are in a hurry, feel free to jump directly in the last wrap-up section).
Installing RStudio server
Trying to install the RStudio server using the provided script install_rstudio.sh, I faced the following error:
[oracle@bigdatalite ~]$ cd scripts [oracle@bigdatalite scripts]$ install_rstudio.sh Retrieving RStudio --2015-10-14 08:01:55-- http://download2.rstudio.org/rstudio-server-0.98.1062-x86_64.rpm Resolving www-proxy.us.oracle.com... failed: Name or service not known. wget: unable to resolve host address “www-proxy.us.oracle.com” Installing RStudio Loaded plugins: refresh-packagekit, security Setting up Install Process public_ol6_latest | 1.4 kB 00:00 public_ol6_latest/primary | 53 MB 01:18 public_ol6_latest 32348/32348 No package rstudio-server-0.98.1062-x86_64.rpm available. Error: Nothing to do cp: cannot create regular file `/etc/rstudio/': Is a directory Restarting RStudio sudo: /usr/lib/rstudio-server/bin/rstudio-server: command not found sudo: /usr/lib/rstudio-server/bin/rstudio-server: command not found
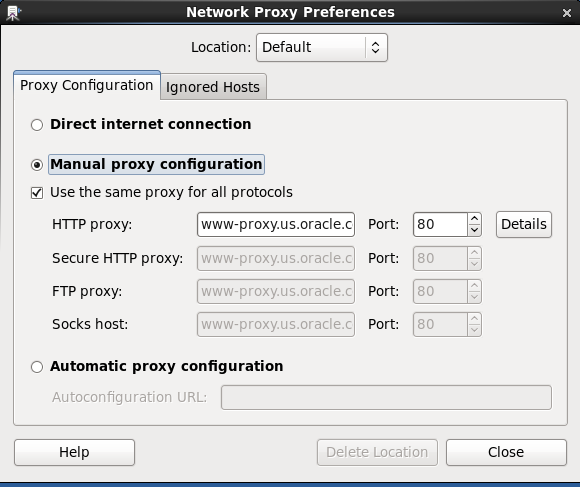
Consulting with our Linux expert sysadmin, Chris Vezalis, and judging from the message Resolving www-proxy.us.oracle.com... failed: Name or service not known, it turned out that a manual proxy has been configured in the VM; from the VM menu, select System -> Preferences -> Network Proxy:
Selecting “Direct internet connection” in the screen above, the installation proceeds without a problem.
On a side note, it is not clear to me why Oracle insists in using this particular version of RStudio server (0.98.1062), which is now more than a year old and superseded by 15 latest releases (see here); in case you want to use the latest version of RStudio server (0.99.486 as of October 7, 2015), edit the install_rstudio.sh script by replacing the wget and sudo yum install commands with the following ones:
wget https://download2.rstudio.org/rstudio-server-rhel-0.99.486-x86_64.rpm --header "Referer: download2.rstudio.org" sudo yum install --nogpgcheck rstudio-server-rhel-0.99.486-x86_64.rpm
Install additional R packages
Most of the additional R packages are successfully installed, with two exceptions; the first is arulesViz, which needs a more recent version of the arules package than the one already installed:
[oracle@bigdatalite scripts]$ install_additional_packages.sh [...] Error : package ‘arules’ 1.1-3 was found, but >= 1.2.0 is required by ‘arulesViz’ ERROR: lazy loading failed for package ‘arulesViz’ * removing ‘/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library/arulesViz’ [...]
We have also the minor (and expected) issue of some dependent packages that reside in Bioconductor (instead of CRAN) reported as “not available”:
Warning: dependency ‘graph’ is not available # for igraph Warning: dependencies ‘graph’, ‘Rgraphviz’ are not available # for arulesViz Warning: dependency ‘highlight’ is not available # for Rcpp
Unfortunately, trying to update arules to a more recent version also fails:
[oracle@bigdatalite ~]$ Rscript --verbose -e 'install.packages("arules",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")'
[...]
** preparing package for lazy loading
Error in matrix(ncol = 0, nrow = nrow(.Object)) :
non-numeric matrix extent
Error : unable to load R code in package ‘arules’
ERROR: lazy loading failed for package ‘arules’
* removing ‘/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library/arules’
* restoring previous ‘/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library/arules
I strongly suspect that this is an issue of the package itself; I also tried to manually download and install both the previous stable version from CRAN (1.2-0), as well as the latest development version from R-Forge (1.2-1.1), in vein.
What we can do is download manually and install a previous version of arulesViz, which does not depend on the most recent version of arules; and this turns out to work for arulesViz version 1.0-0:
[oracle@bigdatalite ~]$ wget https://cran.r-project.org/src/contrib/Archive/arulesViz/arulesViz_1.0-0.tar.gz
[...]
Saving to: “arulesViz_1.0-0.tar.gz”
[oracle@bigdatalite ~]$ Rscript --verbose -e 'install.packages("arulesViz_1.0-0.tar.gz",repos=NULL,dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")'
[...]
* DONE (arulesViz)
[oracle@bigdatalite ~]$ rm arules*
The second package that fails to install is iplots, which is a dependency of arulesViz:
** preparing package for lazy loading Error : .onLoad failed in loadNamespace() for 'rJava', details: call: dyn.load(file, DLLpath = DLLpath, ...) error: unable to load shared object '/usr/lib64/R/library/rJava/libs/rJava.so': libjvm.so: cannot open shared object file: No such file or directory Error : package ‘rJava’ could not be loaded ERROR: lazy loading failed for package ‘iplots’ * removing ‘/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library/iplots’
The reason for this is that, as we have remarked in the past, the command sudo R CMD javareconf, issued in the beginning of the install_additional_packages.sh script, is not enough to fully reconfigure Java for R; it needs an additional flag -E:
[oracle@bigdatalite ~]$ sudo -E R CMD javareconf
[...]
[oracle@bigdatalite ~]$ Rscript --verbose -e 'install.packages("iplots",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")'
[...]
** testing if installed package can be loaded
* DONE (iplots)
A final comment would be that, while the script explicitly installs the packages Rcpp and colorspace, this is not necessary, as the subject packages have already been installed as dependencies of igraph, hence the relevant lines in the script can be commented out.
Install more packages from the R shell
Before proceeding here, let me point out that, if you start R from the shell and execute the .libPaths command before installing the RStudio server, you will get what is shown below:
> .libPaths() # BEFORE RStudio installation [1] "/usr/lib64/R/library" [2] "/usr/share/R/library" [3] "/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library"
This is in good agreement with what the case was in previous versions of the VM, e.g. in version 4.1:
> .libPaths() # in VM 4.1 [1] "/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library" [2] "/usr/lib64/R/library" [3] "/usr/share/R/library"
(If you see a small and seemingly innocent difference, keep a notice; we will need it later).
If we run the same command now, after we have installed the RSudio server as explained above, we get the following:
> .libPaths() # AFTER RStudio installation [1] "/home/oracle/R/x86_64-unknown-linux-gnu-library/3.1" [2] "/usr/lib64/R/library" [3] "/usr/share/R/library" [4] "/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library"
What has happened? Well, RStudio tried to find a location to put its own two packages, rstudio and manipulate; and since the first location listed in .libPaths, /usr/lib64/R/library, is not writable (more on this in a second), it created a new “personal” library directory, using the default location shown first in the list above. Indeed, we now have one more directory named R in our home folder, not present before:
[oracle@bigdatalite ~]$ ls Desktop Downloads movie oradiag_oracle Public scripts Videos Documents GettingStarted Music Pictures R Templates
Might be just a matter of taste, but this is highly undesirable: having already 3 library locations, we would certainly not like a fourth one; moreover (and most importantly), with this library path structure, all packages we will install in the future by simply calling install.packages('package_name') (i.e. without specifying a location) will by default be written in this “spurious” directory.
So, the suggested action is to move these two RStudio packages in our “main” library location, and then delete this new R directory, as follows:
[oracle@bigdatalite ~]$ mv /home/oracle/R/x86_64-unknown-linux-gnu-library/3.1/* /u01/app/oracle/product/12.1.0.2/dbhome_1/R/library [oracle@bigdatalite ~]$ ls /u01/app/oracle/product/12.1.0.2/dbhome_1/R/library acepack forecast iterators ORCHtestkit png seriation ape Formula its ORE praise statmod arules fpp kernlab OREbase proto stringi arulesViz fracdiff labeling OREcommon quadprog stringr bitops gclus latticeExtra OREdm rbenchmark testthat Cairo gdata lmtest OREeda RColorBrewer timeDate caTools ggplot2 longmemo OREembed Rcpp tseries colorspace gplots magrittr OREgraphics RcppArmadillo TSP crayon gridBase manipulate OREmodels registry urca date gridExtra memoise OREpredict reshape2 vcd DBI gtable munsell OREserver rgl XML dichromat gtools mvtnorm OREstats rngtools xtable digest Hmisc NMF ORExml ROracle zoo doParallel igraph nnet pkgKitten rstudio expsmooth igraphdata ORCH pkgmaker RUnit fma inline ORCHcore plyr scales foreach irlba ORCHstats pmml scatterplot3d [oracle@bigdatalite ~]$ rm -r /home/oracle/R
The two RStudio packages, rstudio and manipulate, have been transferred to our “main” user library (see highlighted lines); and if we check with .libPaths again, we will see that indeed the “spurious” directory has gone (not shown here).
So, let’s try now to install more packages from the R shell:
> install.packages('gbm')
Installing package into ‘/usr/lib64/R/library’
(as ‘lib’ is unspecified)
Warning in install.packages("gbm") :
'lib = "/usr/lib64/R/library"' is not writable
Would you like to use a personal library instead? (y/n) n
Error in install.packages("gbm") : unable to install packages
We answered no when prompted if we want a personal library (highlighted line above), since, as explained above, we have already 3 library locations and we wouldn’t want to add a 4th one.
What happens is that the install.packages function, if not provided with a specific library location, tries to install the packages in the first location as listed by .libPaths above; and, once the first location is not writable, it offers to create a new location.
What we have to do is simply to change the ordering of the listed locations, as follows:
> new <- c("/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library", "/usr/lib64/R/library", "/usr/share/R/library") > .libPaths(new)
> .libPaths()
[1] "/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library"
[2] "/usr/lib64/R/library"
[3] "/usr/share/R/library"
> install.packages('gbm')
Installing package into ‘/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library’
(as ‘lib’ is unspecified)
--- Please select a CRAN mirror for use in this session ---
[...]
** building package indices
** testing if installed package can be loaded
* DONE (gbm)
The above changes in the library paths are temporary; in order to make them permanent, open the .Rprofile file in the home directory (gedit ~/.Rprofile), and change it as follows (it currently contains only a commented-out line):
#.libPaths("/u01/app/oracle/product/12.1.0/dbhome_1/R/library")
.libPaths(c("/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library", "/usr/lib64/R/library", "/usr/share/R/library") )
Wrap-up (and the correct order of actions)
In summary, here is the correct order one should perform the required actions in order to come up with a working installation of R & RStudio:
- Open System -> Preferences -> Network Proxy, and select “Direct internet connection”
- Edit your
~/.Rprofilefile, by adding the highlighted line below:#.libPaths("/u01/app/oracle/product/12.1.0/dbhome_1/R/library") .libPaths(c("/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library", "/usr/lib64/R/library", "/usr/share/R/library") ) - Run
~/scripts/install_rstudio.sh - Edit
~/scripts/install_additional_packages.shas shown in the highlighted lines below:# Install additional open-source R packages for HOL exercises # Main packages are arules, arulesViz and forecast plus their dependencies # export http_proxy=http://www-proxy.us.oracle.com:80 echo Configuring JAVA Environment for R sudo -E R CMD javareconf echo Installing additional packages Rscript --verbose -e 'install.packages("igraph",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' Rscript --verbose -e 'install.packages("arulesViz",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' Rscript --verbose -e 'install.packages("tseries",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' Rscript --verbose -e 'install.packages("fracdiff",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' # Rscript --verbose -e 'install.packages("Rcpp",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' Rscript --verbose -e 'install.packages("RcppArmadillo",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' Rscript --verbose -e 'install.packages("nnet",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' # Rscript --verbose -e 'install.packages("colorspace",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' Rscript --verbose -e 'install.packages("timeDate",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' Rscript --verbose -e 'install.packages("forecast",repos="http://cran.us.r-project.org",dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' - Run
~/scripts/install_additional_packages.sh - Run the following commands from the shell, so as to get an older (albeit functional) version of package
arulesViz:wget https://cran.r-project.org/src/contrib/Archive/arulesViz/arulesViz_1.0-0.tar.gz Rscript --verbose -e 'install.packages("arulesViz_1.0-0.tar.gz",repos=NULL,dependencies=TRUE,lib="/u01/app/oracle/product/12.1.0.2/dbhome_1/R/library")' rm arules*
and you should be set!
And just in case you are also going to use Oracle Big Data Discovery in the VM, be sure to check this post too for a configuration issue.-
- Streaming data from Raspberry Pi to Oracle NoSQL via Node-RED - February 13, 2017
- Dynamically switch Keras backend in Jupyter notebooks - January 10, 2017
- sparklyr: a test drive on YARN - November 7, 2016